Research activities

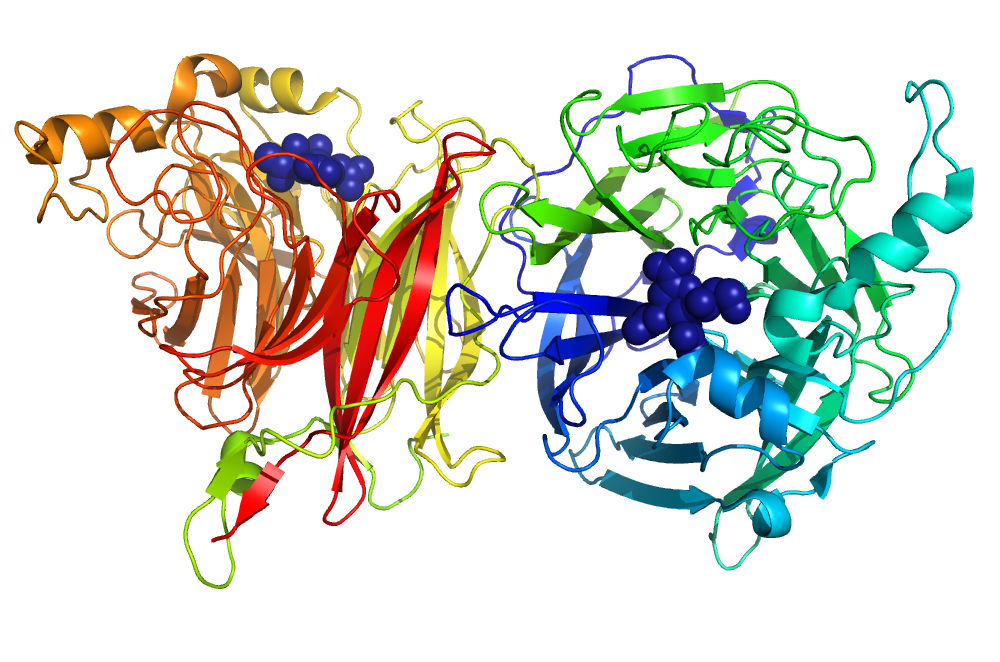
We initially identify new host-pathogen and host-host protein interactions using a proteomics approach. We then explore the significance of these novel interactions using a combined structural (e.g. X-ray crystallography, SAXS), biophysical (e.g. analytical ultracentrifugation) and functional approach (e.g in vitro assays, cell-based assays). Pathogens of interest include influenza, HIV, respiratory syncytial virus, Zika virus, dengue virus, West Nile virus and members of the Enterobacteriaceae (eg. Salmonella Shigella).
With drug-resistance rising, the additional absence of therapies for many existing viral/bacterial infections, and the rise in the number of emerging viral pathogens, the WHO warns of an impending global crisis that will threaten human health and the global economy on a scale even beyond the current impact of the COVID-19 pandemic.
Host organisms detect and clear infection by coordinating between the innate and adaptive arms of immunity. We’re interested in innate host defences triggered by the NOD-like receptor 3 (NLRP3) inflammasome, which responds to microbial and danger signals to produce proinflammatory cytokines that can lead to inflammatory cell death. We also study the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), which act as sentinels for viral RNA from pathogens of significance to human health such as COVID-19, influenza, hepatitis B, hepatitis C and respiratory syncytial virus.
We are interested in identifying new connections between proteins in these innate immune signalling cascades and the functional relevance of the interactions.
The success of a pathogen is partly determined by how well it can utilise host cell proteins for replication and also evade the host innate immune response. Well-known RNA viruses like influenza A and hepatitis C implement immune evasion strategies. Unfortunately, these immune evasion mechanisms are often poorly understood and hampered by our limited understanding of how antiviral immunity is activated by innate immune receptors such RIG-I. We are interested in mechanistically understanding how microbial pathogens target host cell proteins within the RLR signalling cascade.
The correct subcellular localisation of host and microbial proteins is central to their function. We are interested in better understanding how host and microbial proteins use nuclear transport receptors to shuttle between the nucleus and cytoplasm. Agents that selectively alter the nuclear transport of host or microbial proteins have potential application as therapeutics for microbial pathogens of significance.
We use a combined approach to study our innate immune response to infection. Techniques used within the laboratory include molecular biology, recombinant protein expression, protein purification, X-ray crystallography, small angle X-ray scattering, analytical ultracentrifugation, biochemical assays, high-resolution imaging, gene knockdown/knockouts, luciferase assays, coimmunoprecipitation.
Acknowledgement of Country
RMIT University acknowledges the people of the Woi wurrung and Boon wurrung language groups of the eastern Kulin Nation on whose unceded lands we conduct the business of the University. RMIT University respectfully acknowledges their Ancestors and Elders, past and present. RMIT also acknowledges the Traditional Custodians and their Ancestors of the lands and waters across Australia where we conduct our business - Artwork 'Luwaytini' by Mark Cleaver, Palawa.
Acknowledgement of Country
RMIT University acknowledges the people of the Woi wurrung and Boon wurrung language groups of the eastern Kulin Nation on whose unceded lands we conduct the business of the University. RMIT University respectfully acknowledges their Ancestors and Elders, past and present. RMIT also acknowledges the Traditional Custodians and their Ancestors of the lands and waters across Australia where we conduct our business.